We Designed the Interface
It was clear that neuroscientists needed more than merely a database
for performing one-shot queries. In order for the GENESIS database to
be useful it had to provide a means to seamlessly integrate itself
into the larger neuroscience research process. To achieve this,
particular attention was paid to design a tool that hid the fact that
a database was being accessed. Previous database interfaces have
either been too low-level, where the underlying schema is exposed, or
too high-level so that only trivial queries can be made.
Neuroscientists are typically not interested in the underlying data
structure used to store the data. They are more interested in finding
the data and applying it directly to their research tasks. Our
solution, which is an extension of the visualization workspace in our
original proposal, was to design the Neuroscience Information
Workspace which applies the desktop metaphor (popularized on many
graphics workstations today) to manage and organize data extracted
from the database, and to relate it to data typically gathered by
neurobiologists.
Object-Oriented Direct Manipulation
To support the workspace, our design provided a local database to
manage the data that evolves as a result of querying the main database
(meta-data). Interaction with the local database is via an
object-oriented direct-manipulation interface. Every item of data in
the interface is manipulated as an object and each object has its own
interface. This allows us to continuously add new object types to the
system without significantly affecting the system's interface as a
whole. A query agent is introduced to simplify query composition, and
a hierarchical conceptual storage metaphor was included to help users organize
information that they accumulate over time.
Query Agent and Data Orienteering Maps
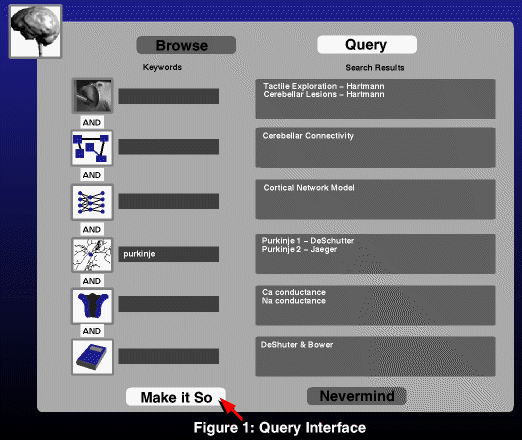
Data is extracted from the main database by two mechanisms: by
activating hypertext links in a document; or by consulting a
context-sensitive query agent (Figure 1). The query agent maintains
information on search paths that have already been traversed
(contexts) so that it can avoid retraversing these paths in answering
follow-up queries (Appendix B) shows a thumbnail sketch of the kinds of
traversals possible with the interface). Each data item extracted is
either placed in, what is called, a temporary notebook for further
exploration; or if the search is a continuation of a search path, will
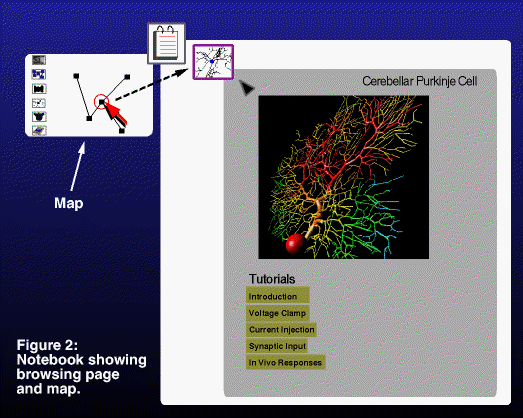
be added as a node in the current notebook. Notebooks provide a means
to view the information sequentially as well as on-demand via a local
search map (Figure 2). Simply put, this map visualizes the
relationships between the search nodes traversed in a session
registered in a notebook. More importantly, when multiple maps are
merged, the result provides a means to relate information between
other notebooks and hence larger contexts. We believe this data
orienteering methodology can provide a powerful means to help
neurobiologists visualize relationships between multiple levels of
neuroscience data. Each local map could represent a subdomain of
interest. A combination of maps could represent the relationships
between the subdomains. We can use this information for two purposes:
to relate information from seemingly disparate research domains; and
to transmit this information back to the main database to augment its
own data store of relationships for the benefit of other database
users.
Information Organization Metaphors
The interface includes a number of mechanisms, borrowed from everyday
familiar objects, for information organization. For example, post-its
are used to attach annotations to arbitrary surfaces; notebooks are
used to quickly collect data; drawers are used as storage for
notebooks. From building our data acquisition prototype (see below),
we discovered that these differing levels of data organization were
needed as a means to enable information sharing between collaborating
scientists.
Support for Multiple Data Granularities; Data Exchange;
and Operators to Process Aggregations of Data
In traditional journal publications, data is provided at one
granularity. Users are then expected to take the data and use whatever
is available to them to extract the fragments they really want. In
fact, from our initial user study, we were quite surprised to discover that in
some instances, neurobiologists would hand-digitize graphs from
journals by using a ruler simply because the numerical form of the
data was not available. We were careful in the design of our
interface, to ensure that data stored in the database could be
retrieved at any granularity. Our object-oriented interface and data
model allows the user to literally "pick up" any piece of data and
export it for use in a number of popular data analysis programs
(Matlab, xplot, gnuplot, etc.) Since our database is heavily
model-based, one of the data formats will encapsulate
GENESIS
simulation models which may be exported to a locally residing
GENESIS
simulator.
In addition to providing such export mechanisms, our interface allows
the user to collect a number of data objects and apply built-in tools
to them. For example, one could collect a number of Purkinje cell
models found on the database, by a number of authors, and apply a
comparison tool to them. This tool would display, in a stereotyped
format, a comparison of the two Purkinje cell implementations- channel
information, morphology, firing data, etc. These tools are maintained
in a toolbox, and new tools can be developed and added with time.
Currently these designs have been sketched as a storyboard. These
will shortly be presented to the neurobiologist for review before a
functional prototype is built. A thumbnail sketch of the storyboard
is included in Appendix B.
We Built a Data Acquisition Prototype
We Identified a Suitable Data Model
Concluding Remarks
 Return to Brain Menu
Return to Brain Menu


