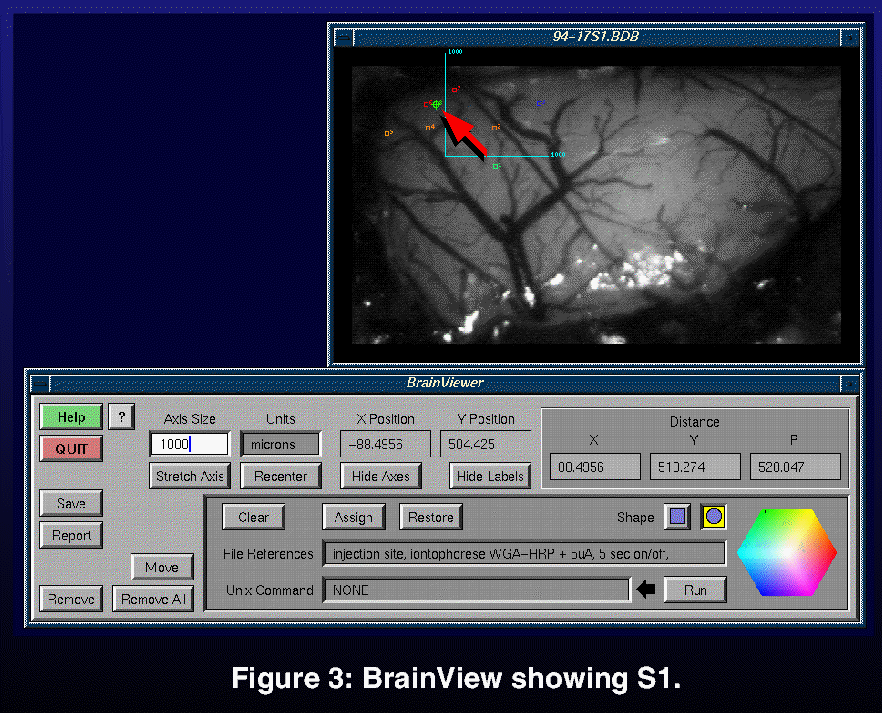
We have built a prototype, called BrainView for data acquisition in mapping experiments (Figure 3). The prototype was written in C++ on a Silicon Graphics (SGI) workstation. A UNIX file-based data store is temporarily being used to delay our having to commit to a particular database model at this early stage of development. The prototype was again motivated by the user study which suggested that there was a real need for an integrated way to 1) store and organize experimental data, 2) to relate this data to model data. Hence users would undoubtedly want to be able to relate their experimental data with data found in the database. This prototype therefore provided a means for us to think more concretely about how we were going to design a data model that could support this connection.

This prototype is currently being used to collect and store electrophysiological micromapping data from S1 of the somatosensory cortex, crusIIa and crusIIb of the cerebellum. The goal there is to precisely determine the anatomical pathways for the circuit between S1 and crusIIa, via the pontine nuclei. It is hoped that by isolating these tracings we can build a connectivity model of these regions of the brain. Use of BrainView involves first digitizing a microscope image of the brain region into the SGI. After digital enhancement, this image is used to create a new data store. BrainView will present this image as part of the database interface, allowing the user to mark electrode penetrations on the image and enter data (mapping notes, digitized physiological data, etc.). This then provides a means, at a later date, to query and further analyse the data. This first design iteration of BrainView was succeeded by a discussion with the neurobiologist who used the system. From this discussion we compiled a list of future extensions to the system.