![\includegraphics[width = 4 in]{top-level.eps}](img7.gif)
|
Michael Hucka, Kavita Shankar, David Beeman
{mhucka,kshankar}@bbb.caltech.edu, dbeeman@dogstar.colorado.edu
Bower Research Laboratory, Division of Biology 216-76
California Institute of Technology, Pasadena, CA 91125
Date: 30 October 2000
The Modeler's Workspace is a software system intended to assist computational neuroscientists in interacting with databases of models and with neural simulation packages such as GENESIS. There are three components in the system: a user interface, a database server, and a global registry and repository.
This document describes the representation of database objects in the Modeler's Workspace. Each object in the system is created according to a specific format. Database designers traditionally call such a format a ``schema'', and object-oriented programmers call it a ``class'', but in an attempt to reduce the amount of abstract jargon in our writing, we simply call it a template. A template defines how a particular kind of database object is to be expressed, meaning the structure of the object's representation, the attributes or slots in the representation, and the types of data values that are permissible for each slot. Different object templates are used to create objects that represent different kinds of information. As explained below, templates in the Modeler's Workspace are organized in a simple hierarchy, and the system provides facilities for definining new templates.
This document is a companion to The Modeler's Workspace Database Organization and Interaction (Hucka, Shankar, and Emardson, 2000). Here we focus on the model representation; in the former, we focus on the relationships and interactions between databases. An overview of the entire system is provided in An Overview of the Modeler's Workspace (Hucka, Shankar, and Emardson, 2000). The database design presented here is based in part on ideas previously developed in versions of the Modeler's Workspace implemented by Jenny Forss, David Beeman, Sara Emardson, and others (Beeman et al., 1997; Forss et al., 1999).
The Modeler's Workspace requires a representation language that abstracts away specifics of particular simulators such as GENESIS and NEURON, as well as provides ways of interacting with existing neuroscience databases on the Internet. Devising such a representation is difficult.
To begin addressing this problem, we first distinguish between a Modeler's Workspace Database, which is the database component of the Modeler's Workspace system, and foreign database, meaning any other kind of database. Since the design and implementation of the former are under our control, a Workspace Database can provide whatever functionality we deem necessary. Foreign databases are outside our control and may not provide the same level of functionality or representational structure.
In order to support some level of interoperability with foreign databases as well as neural simulators, and allow users and software developers to evolve new representations and tools, we use a multifaceted approach having the following key aspects:
The representational framework used in the Modeler's Workspace is based loosely on object-oriented programming concepts. One of the key ideas used is the concept of inheritance applied to templates. All templates are derived from either a specific one called Base or an existing template. Base (defined in Section 3) is therefore the root of a template hierarchy.
![\includegraphics[width = 4 in]{top-level.eps}](img7.gif)
|
The Base template itself contains few attributes, so it is not biased towards any particular kind of database object. It is normally not used to represent objects in and of itself. Figure 1 shows the first level of templates derived from Base in the Modeler's Workspace system. The templates are Author, Reference, Method, Model, Data, and Site; they are defined in detail in Section 3. The particular choice of first-level templates was inspired by the work of Gardner et al. (1999).
Each template inherits the same attributes as the template from which it is derived, and in addition, may add its own set of attributes. Users are allowed to add new attributes when creating a new template based on an existing one, but not to delete attributes. For example, if a given template has attributes x, y and z, a derived template can only add attributes; it cannot change or delete x, y or z. This restriction is necessary to ensure that search operations across databases make sense: if existing template attributes were to be modifiable, there could be no assurance that the fields retained their meanings across different databases, and database communication would become nearly impossible.
The object-oriented style of representation is useful for a variety of reasons. First, the existence of categorical templates allows the Modeler's Workspace User Interface to present the user with intelligent search forms. Specifically, the Modeler's Workspace search interface prompts the user to specify the type of object to search for (which is equivalent to specifying the template), and based on the user's choice, the system constructs a form using knowledge of the attributes defined by the template. The search form may include graphical elements specialized for the particular category of object involved. This allows the system to go beyond the usual fill-in-the-blanks search form and provide something more powerful and user-friendly.
A second reason is that, by chosing the search category appropriately, searches can be made more or less specific. Because of the hierarchical relationships, a user can select a template in the middle levels of the hierarchy, and search operations can be designed to encompass all objects that are below it in the hierarchy. This means, for example, that a search using Model will encompass objects created from templates derived from it, such as Neuron class objects, TransmembraneMechanism class objects, etc.
A final reason for the utility of the representational framework presented here is that software can be made modular and extensible. New software modules can be developed for the Modeler's Workspace alongside new templates, customizing the system to interact with new types of objects without redesigning or restructuring the whole system. For each representation derived from an existing template, all the software elements that worked with the parent template will also work with the derived templates. This is because the derived template can only add attributes, and while the existing tools will ignore the new attributes, they will continue to work with the attributes that were inherited from the parent template. Developers can write new software modules that interact with the additional fields in the new templates and these software modules can be loaded into the Modeler's Workspace on demand, extending the software's functionality.
XML, the Extensible Markup Language (Bosak and Bray, 1999; Bray, Paoli and Sperberg-McQueen, 1998) is a language used to express self-describing, semi-structured representations of information. It provides a way of marking up data with semantic tags that describe and structure the contents of the data. Although XML is typically thought of as a document format similar to HTML, in fact it is more general. It is a notation, a ``metalanguage'', a way of organizing a stream of data and marking up the different parts so that a program can parse the stream into constituents. In the words of one of its chief architects, ``Just as HTML created a way for every computer user to read Internet documents, XML makes it possible, despite the Babel of incompatible computer systems, to create an Esperanto that all can read and write. Unlike most computer data formats, XML markup also makes sense to humans, because it consists of nothing more than ordinary text'' (Bosak and Bray, 1999).
In the Modeler's Workspace, XML Schemas (Biron and Malhotra, 2000; Fallside, 2000; Thompson et al., 2000) are used to describe model templates, and actual models are encoded in XML using these Schemas. All of the constructs described in Sections 3-5 have direct equivalents in XML Schema.
For handling representations in a collection of databases (Modeler's Workspace and foreign databases), XML offers three key features:
The powerful generality of XML suggests that more than just the model representations can be expressed using it, and indeed, the Modeler's Workspace uses XML for all file formats. For example, the Workspace initialization file, storing user's preference settings and other information, is written in an XML format. In addition, glossary and notes are also stored in XML files that are read when the Workspace is started up. Each glossary and note entry has a tag, so that the program can reference each entry directly. This not only makes the information organization cleaner, it also makes it possible to issue updates to the glossary/notes files more easily.
Many of the attributes in the templates have essentially unconstrained values. For example, there are no a priori constraints applicable for such things as the name of a model. But in many other cases it is useful to place constraints on the permissible values of an attribute, especially string attributes, in the form of a controlled vocabulary. Gardner et al. (see http://cortex.med.cornell.edu/dataModel/) are leading an effort to define a set of controlled vocabularies for use in biological databases. These are meant to capture the allowed values for an attribute. A controlled vocabulary is especially useful for search operations: when database objects use attribute values drawn from a common vocabulary, it is much more likely that a database search will succeed in finding a match than if users are given free reign to enter any value for every attribute. Controlled vocabularies reduce the chances that differences in spelling, the use of different terms having the same meaning, and other incidental effects will cause a search to fail.
We borrow this general idea for the Modeler's Workspace, but in some situations we also allow for attribute values that are not strictly limited to values from a set vocabulary. We call this a semi-controlled vocabulary. Such semi-controlled vocabularies can be attached to an attribute to provide a set of initial, suggested values. When these are available for a given attribute in a model template, the editing field for that attribute provides a pull-down list containing the set of suggested values, allowing the user to easily select a value from the list. In addition, the user is allowed to type in a new value if none of the existing values are suitable.
The use of a semi-controlled vocabulary involves a tradeoff as compared to a strict controlled vocabulary. The provision for allowing users to type in new values means that search operations may no longer be as effective. However, we feel that we are not able to define a sufficiently comprehensive controlled vocabulary for all attributes, and that moreover, users would react negatively to an interface that does not allow them to type in new attribute values when the predefined set is insufficient. We believe that we can obtain most of the benefits of a controlled-vocabulary approach by providing reasonably comprehensive default vocabularies, so that users are likely to find a predefined value close enough for their needs.
The graphical notation for defining templates that we use throughout this document is explained separately in The Modeler's Workspace Notation for Describing Representations Intended for XML Encoding (Hucka, 2000). We urge readers to peruse this document before proceeding further here.
The six most basic templates derived from Base, shown in Figure 1, are Author, Reference, Method, Model, Data, and Site. These are general data structures that are not limited to representing specific biological objects such as neurons and ion channels.
In this section, we detail the preliminary versions of the six basic templates. Some of them, such as Author and Reference, can be used directly, and we expect that they will not require further specialization. Others, such as Model, clearly need to be specialized to apply them to particular areas. The specializations that we have developed so far for the Modeler's Workspace are intended for representing models of single neurons and associated elements such as ion channels. Sections 4 and 5 describe the templates for models of neuronal cells and various cell mechanisms.
The Base template has only two attributes, id and version. The former places a unique identifier on every object; the latter allows the system to track the evolution of data objects. The diagram below depicts the Base template and the Version structure:
![\includegraphics[scale = 0.65]{basetemplate.eps}](img8.gif)
The id attribute of type MWSUID is an identifier that is unique to a given Workspace Database. Every object is given a unique identifier so that other objects can refer to it. For example, a model of a neuron may refer to several models of ion channels as part of its definition. In order for the system to be able to link models together, the database objects themselves must have identifiers that are unique within a given Workspace Database server. This identifier must be recorded in the database object itself. The name of an object (for those database objects that have names, such as Model, described in Section 3.5) is not sufficient in this capacity because the name can be changed by the user. The procedure used in the Modeler's Workspace system for generating unique identifiers is described in Appendix A.
The Version structure for attribute version contains fields for such things as a timestamp, a version number, the version number of the immediate parent version, etc. Two of the attributes are required to be given values: timeStamp, the time when the data object being described was stored (represented using the XML Schema datatype timeInstant); and versionNumber, a descriptor for the particular version of the data object being described. The remaining information in the Version structure is optional: versionName, a symbolic name or tag for the version (e.g., ``RELEASE_1''); logMessage, the user's description of what has been changed in a given version; parentVersion, the version that is the ancestor to the current one; and source, a reference to the object in question. Despite their names, the version ``numbers'' are actually stored as strings, because in many version control systems such as RCS and CVS (Fogel, 1999; Mikkelsen and Pheriogo, 1997), version numbers have the form ``1.4.9'' and these cannot be represented as integer or floating point quantities. The source attribute allows an object to retain a reference to the original database object from which it came, so that when copies of the object are made and communicated on a network, they retain information about the original source. (This is analogous to the $Source$ keyword in RCS/CVS.)
Since all objects must be derived from Base, all objects inherit id and version attributes. This may at first seem odd, because it may seem that some types of objects such as ``data'' do not need versions. However, we believe that all database object representations can benefit from having version information, because it enables changes (such as the addition of more details) to be tracked through a version control system. The Modeler's Workspace will include version control facilities for managing models in a Workspace Database.
![\includegraphics[scale = 0.65]{authortemplate.eps}](img9.gif)
The Author template inherits the id and version attributes from the Base template, and adds attributes for identifying a person by name, address, web home page and other characteristics, as shown in the diagram at the right. (The attributes implicitly inherited from the Base template are not repeated here.)
The primary purpose of a separate Author template is to allow users to enter into their databases the information about a given author once, then link to the author information from other objects (such as models and article references). This will produce savings in effort as well as storage space. This will also permit the construction of databases devoted to author information. We envision that such databases could be constructed using, for example, the author information contained in the proceedings from the annual Computational Neuroscience conference, and then made available as a public resource on the Internet. Users of the Modeler's Workspace could then import author records from this database server, potentially avoiding having to enter author information altogether.
The following is an example of a data object encoded in XML, based on the
template defined above and using the UML-to-XML translation approach
described by Hucka (2000):

This example illustrates several points. The first line begins with the tag mwml, indicating that the rest of the data stream is encoded using the Modeler's Workspace Markup Language, an application of XML. MWML is defined in Section B. The first line also contains a version number, indicating that the data stream is based on version 1 of the MWML specification. (Such versioning allows for future updates to MWML.) Subsequent lines contain the definition of an author object, with a variety of XML attributes encoding information about the author. The value of id is artificial and shown for illustrative purposes only. The version information, because it is defined as a substructure (see the definition of Base in the previous section), is structured as a subelement inside author, with the tag version. Both the author and version elements omit mention of certain optional attributes that have not been given values in this example.
![\includegraphics[scale = 0.63]{referencetemplate.eps}](img11.gif)
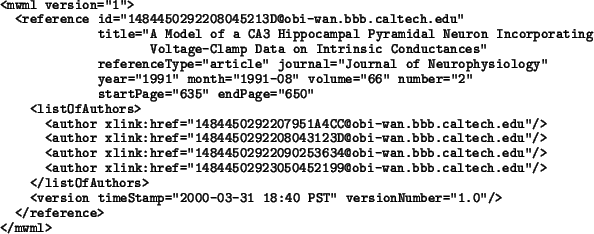
Not all of the attributes in Reference will be used in every literature reference. For any given Reference object, attribute referenceType indicates the kind of reference it is, whether a book, journal article, etc. In a well-designed user interface, the choice of referenceType made by the user can trigger the interface to selectively present only those attributes relevant to the type of reference in question. Thus, if the user is creating an article reference, attributes such as journal and month would be significant and worth showing in the interface, but if the user is creating a book reference, these attributes would not be shown; instead, other attributes such as chapter and editor would be relevant.
As with the Author template described above, the use of a separate Reference template will allow users to enter common literature information into their databases and reuse them when creating models. We also envision that databases containing reference information could be established as public resources on the Internet. Their contents could be created automatically using software that converts information from PubMed and other sources.
The following is an example article reference encoded in XML. In this
example, the author information is shown as a list of references following
an XML XLink syntax. At this time, the exact form of the URIs for linking
to database objects is unknown, so in this example, the four authors of the
paper are shown as links to imaginary database objects named
1484450292207951A4CC@obi-wan.bbb.caltech.edu, etc. The
reference are all assumed to point to objects in the same Workspace
Database. (The object references may be different in actual database
implementations.)
The Data template is another subclass of Base. It provides basic support for storing data in a workspace database or pointing to data stored in a remote database.
![\includegraphics[scale = 0.65]{datatemplate.eps}](img13.gif)
The title and description attributes can be used to provide information about the data for human readers. The author list is intended to point to the ``authors'' of the data, meaning the individuals responsible for gathering or otherwise producing the data. The reference list is intended to point to relevant literature references; for example, if the database object is an electronic form of a set of data published in a journal article, one of the references should point to a Reference database object that provides information about the article.
The dataView attribute is a list of DataView structures that can be used to point to data sets. A DataView contains attributes such as viewLabel, viewSequence, and dateAndTime, that allow users to describe a particular view of a data set. The actual data set is stored on in the attribute dataSet, a list of DataSet structures.
A DataSet structure contains a number of attributes:
Note that since both the dataView and dataSet attributes are lists, a given object can have more than one DataView structure, each containing more than one DataSet, leading to a flexible organization of information. The Data template and this arrangement of attributes is based partly on Gardner et al.'s (1999) Data_Element structure.
The following is an example of how an object created according to
Data might appear in the form of an XML data stream:
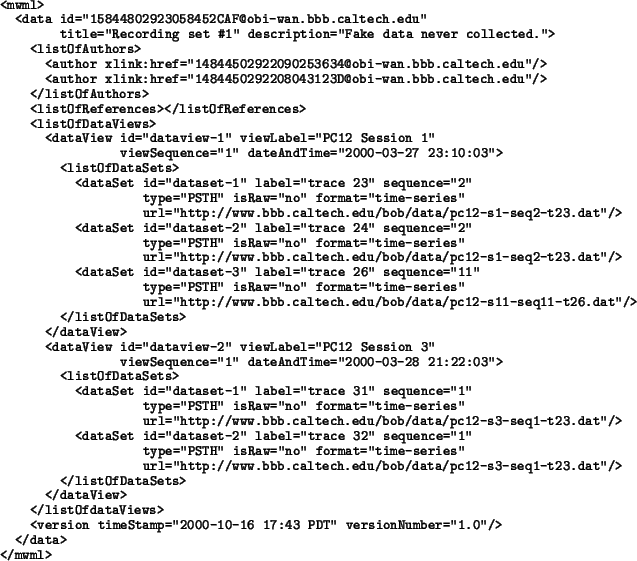
The example shows a data object entitled ``Recording set #1", with two authors (expressed as links to other objects presumably located in the same database). There are no links to Reference items. This data object has two DataView structures, the first of which having three DataSet structures and the second having two. All of the data sets are references to external data located at particular URLs on the Internet.
In the Modeler's Workspace, the primary reason for providing a separate class of objects for storing information about experimental data is that, generally, many models may be based on the same data sets. Therefore, it is useful to be able to store information about experimental data as an independent entity.
The Model template is intended to serve as a common starting point for all model template definitions. It is a generic structure, not specific to any particular kind of modeling. Specific kinds of models, such as for neuronal cells (Section 4) and intracellular and transmembrane mechanisms (Section 5), are derived by starting from Model and adding new attributes.
![\includegraphics[scale = 0.65]{modeltemplate.eps}](img15.gif)
As with the other main templates, the Model template is an extension of the Base template and therefore implicitly inherits id and version attributes. Model then adds several more attributes. The attributes name and description allow a user to name a model and provide a brief description of it. To provide for the possibility of including equations, superscripts, and other formatted content, the description is stored in XHTML format. The notes attribute provides a place for recording information about a model that is not easily recorded in any other attribute. One example of the use of notes is to paste plot output resulting from simulating the model; a model's author might choose to include such plot output for the benefit of other researchers.
The lists for author and reference information serve the same roles as they do in other templates. The author list is intended to point to the ``authors'' of the model; the reference list is intended to point to relevant literature, specifically articles discussing the model. For example, if a database object is related to a model published in a journal article, one of the references should point to a Reference object that provides information about the article. It is important to note, however, that the author list should not point to the authors of the article, unless the authors of the article are also the authors of the model definition.
The version attribute inherited from Base is especially useful in the context of models, because it allows the Modeler's Workspace database to track the history of modifications to user's models.
The Method template is intended to capture information about experimental methodologies. The Site template is intended to capture information about such things as neuronal recording sites, brain regions, etc. These templates have not yet been defined and await further development.
The main template for representing models of neurons is Neuron. It makes use of several other templates, in particular NeuronalAnatomy, NeuronPart, TransmembraneMechanism and IntracellularMechanism. Figure 2 depicts the hierarchical relationships between the templates. In this section, we discuss the Neuron, NeuronalAnatomy and NeuronPart templates. We leave the cell mechanism templates to Section 5.
![\includegraphics[scale = 0.58]{top-level-full.eps}](img16.gif)
|
NeuronalAnatomy specializes the basic Data template to provide a container for anatomical information about neurons. It is primarily intended to be used for storing information about cell morphologies.
![\includegraphics[scale = 0.65]{neuronalanatomytemplate.eps}](img17.gif)
As the NeuronalAnatomy template inherits from Data, it has the data containers defined by the parent template. In particular, NeuronalAnatomy inherits a dataView list attribute that can be used to store a list of either references to or actual data describing the morphology of a neuron. Attribute neuronType is a string that is intended to allow users to describe the type of neuron involved. An example value might be ``pyramidal''. The value supplied by the user is entirely their choice. The attribute cellLocation is a string for describing the anatomical location of the neuron. The attributes neurotransmitters and neuroreceptors are intended to summarize the neurotransmitters with which the neuron may interact and the neuroreceptors to which the neuron may respond. They are lists of strings, with each string naming one substance. A neuron that is sensitive to five neurotransmitters would have five strings in the attribute neurotransmitters. The cellLocation, neurotransmitters and neuroreceptors attributes have semi-controlled vocabularies associated with them, following the approach taken by Gardner et al. (1999).
The main purpose of the semi-controlled vocabularies for the Modeler's Workspace is to allow the system to provide, in the user interface, an initial list of possible values as hints for the user. We believe this will both help users enter information into models and also increase the chances that different models are more likely to use consistent nomenclature. The semi-controlled vocabularies will be hierarchical in nature. Figure 3 lists the current vocabularies for the attributes cellLocation, neurotransmitters and neuroreceptors.
Information about the subject from which the anatomical data is drawn can be recorded in the optional subject field. The associated Subject structure provides slots for a description of the kind of subject (e.g., ``rat''), whether it's male or female, and its age. The age attribute has a string data type rather than a numerical type because users may need the freedom of expressing age in a variety of forms, such as ``1 year 2 months'' or ``4 hours''.
|
The Neuron template extends the basic Model template with additional attributes for anatomical information, experimental information, the segment structure of the model neuron, and other characteristics. Neuron is fairly complex. We begin by discussing the basic idea behind its form.
![\includegraphics[scale = 0.65]{neuronmodel2.eps}](img18.gif)
In GENESIS, models of neurons are described in terms of compartments, whereas in NEURON, models are described using the higher-level construct of cable segments. One of the parameters in a NEURON model determines the number of compartments that a given segment should be divided into at simulation time. We adopt NEURON's latter approach here, and describe neurons fundamentally in terms of segments. This allows a model to be mapped to either GENESIS or NEURON; to translate models into GENESIS, the Modeler's Workspace simply needs to pre-divide the segment structure into compartments and then send the compartmentalized version to GENESIS. The reverse mapping, from a GENESIS model to a model in the current framework, can be implemented in the worst case by treating each GENESIS compartment as one segment. More realistically, however, we believe the system will be able to employ some heuristics in many cases to map multiple compartments into single segments.
In order to support the ability to define entire portions of neurons as building-block parts, and thereby have the ability to create and maintain parts libraries, the representational framework shown here introduces one more intermediate level of abstraction, the section. A section in a Neuron model is either a segment or a reference to a NeuronPart object (see Section 4.3). A NeuronPart object is simply a collection of segments that define some useful building-block structure. A model neuron is defined in terms of one or more sections, each of which is a membrane segment or a reference to a separate part model object.
The Section structure shown in the diagram of Neuron is
an abstract type; the structures MembraneSegment and
PartReference are derived from Section and are the actual
types used for the section list in a Neuron object.
MembraneSegment is used to define a segment directly;
PartReference is (as its name implies) a reference to a collection
of segments designated as a part and stored as a separate database object.
In XML terms, the structure of a model is along the lines of the following
partial example:

In the fictitious example above, most attributes are omitted for clarity.
The definition of the model is in terms of six elements listed under
section. Note how segment definitions are interspersed with part
references. The XML attribute xsi:type is used to specify whether
a given section element is of the derived type
MembraneSegment or PartReference, in accordance with the
XML Schema standard involving the use of derived types (Fallside, 2000).
The id identifiers are numbered consecutively for each section in
the section list, whether a given section is a segment or a part
reference.
The following drawing illustrates how a model can be structured in terms of segments and part references. This uses the same components as the example above:
![\includegraphics[scale = 0.68]{part-example.eps}](img20.gif)
We now turn to describing the different attributes and substructures in the Neuron template. At the highest level, there are are several general attributes. The attributes temperature, Q10Factor and scalingFactor specify the temperature assumed for the parameter values in the model, and scaling factors to be used to adjust parameters for other temperatures if needed. The neuronanatomy attribute in Neuron is a link to an object based on NeuronalAnatomy. The intent is to allow modelers to store or point to the experimentally-derived cell morphology upon which a given neuron model is based. Of course, a given model may not have an associated experimental morphology, in which case, the neuroanatomy attribute would have an empty value. The experiments attribute is a list of links to Data objects. Its purpose is to give users the ability to store links to experimental data about the neuron they are modeling.
Both MembraneSegment and PartReference inherit four attributes from the definition of Section. Attribute id is a unique identifier for the section. Attribute parent is the value of the id of the parent section to which a given section is connected. Attribute name is intended to allow the user to give a name to a segment for their own use; similarly, attribute type is intended to allow a user to describe the kind of anatomical structure of which the segment is a part. For example, in a model containing an apical dendrite described using a number of segments, a user might give the dendritic segments the names ``dend1'', ``dend2'', etc., and assign the string ``apical dendrite'' as the type of all the segments describing the dendrite. These two attributes are entirely for the user's benefit and do not affect how a model is translated into a particular simulator's scripting language; the user could just as well assign gibberish or no value at all to the name and type attributes of all the segments in their model.
The geometry of a segment is described using a collection of attributes gathered under a SegmentGeometry structure. For a given segment, the numberOfCompartments attribute indicates to a simulator such as GENESIS or NEURON the number of compartments into which the segment should be divided. The boolean attribute symmetrical determines whether the compartments should be made symmetrical with respect to the placement of the axial resistance; this attribute is primarily useful for simulators such as GENESIS that make a distinction between symmetrical and asymmetrical compartments. The attribute absoluteDimensions indicates whether the geometry values are measured relative to a parent segment or are given as absolute values. The attribute dimensionUnits specifies the units assumed for the coordinate and dimension attributes. The attributes x, y, and z indicate the dimensions of the segment, while the length attribute sets the length of the segment. The shape attribute specifies the shape of the segment; currently the only two values allowed are ``cylindrical'' and ``spherical'', after the same options available in GENESIS for compartment shapes.
The startDiameter and endDiameter attributes indicate the diameters of the two ends of the segment cylinder. The two diameters allow for segments that are tapered (i.e., cones). For simulators that can handle conically-shaped compartments, the two diameters can be used to calculate the starting and ending diameters of each compartment that make up the segment of membrane; for simulators that cannot handle conical compartments, the segment can be turned into a series of cylindrical compartments, each of which is of a different diameter than the previous one, thereby simulating a tapering segment.
The passive properties of a membrane segment are set using the attributes in the PassiveProperties structure. These are all the common variables used in simulating neuronal membranes in GENESIS and NEURON: specificResistance, specificCapacitance, specificAxialResistance, membraneRestingPotential, membraneLeakagePotential, equilibriumPotential, and maxElectrotonicLength. Each of these attributes also has an associated _units attribute that specifies the units of the value.
A membrane segment can have zero or more active properties identified by a structure of type ActiveProperty. An active property can be an ion channel or other kind of intracellular or transmembrane mechanism. An ActiveProperty structure only contains information linking to another database object and a set of variable assignments. The assumption is that channels and other mechanisms are complex enough to require their own separate object representations. The assign attribute is a two-dimensional list of strings that is intended to let a user specify values for variables in the pointed-to model. The idea is to allow users to put variables in places where quantities are required in a mechanism model, and then use the variableAssignments attribute to tell the simulation program what values to give those variables.
Finally, the messages attribute in MembraneSegment is used to tie together elements that need to exchange parameter values, for example for calcium concentration. The messages attribute is a list of zero or more Message structures. The sources and destinations of messages can be other segments; they can also be active property components such as channels. The sources and destinations are specified in terms of identifiers; since every segment and active property structure has a unique identifier, message can be unambiguously directed to/from any other active property or segment within a model. Each message has a messageName, which is used to specify the name of a variable that is to be exchanged between the destination and target elements.
As mentioned above, a section of a neuron model can also be specified as a reference to a part model. This is handled using a PartReference data structure. Like the Segment structure, the PartReference structure is a subclass of Section, which means it inherits attributes id, name, type, and parent. In addition, the part structure adds a link to a NeuronPart model object. It also adds attributes attachmentPoint (which indictates how far from the end point of the parent segment the part should be attached) and orientation (which indicates the three-dimensional orientation of the part with respect to the parent segment). The assignn attribute is used to specify how variables in the part model should be given values.
The NeuronPart template exists to provide a way to construct reusable part models for Neuron objects. Portions of neuron models can be recorded in NeuronPart objects, allowing those portions to be reused in models by linking to them from within Neuron objects.
The body of a NeuronPart object consists of a list of MembraneSegment structures and a separate list of VariableAssignment structures. The definitions of these structures are taken directly from the structures used in the Neuron template.
![\includegraphics[scale = 0.65]{neuronparttemplate.eps}](img21.gif)
The list of MembraneSegment elements is used to define the part in terms of membrane segments. The segments can have all the attributes of segments in full Neuron objects; for example, they can refer to active properties such as ion channels.
The attribute assign holds a list of VariableAssignment structures. These are intended to support the use of variables as substitutes for numerical or other values in attributes. The additional capability of using variables in place of actual attribute values is necessary to permit part models to be parametrized. The variables can be given values in the Neuron object that links to them, as described in the previous section.
The templates described in this section are used to represent active properties in neural cell models. We begin by describing the TransmembraneMechanism template and its subclasses, and then describe the IntracellularMechanism template and its subclasses beginning in Section 5.3.
The TransmembraneMechanism template is intended to serve as a starting point for definitions of cell mechanisms such as ion channels, calcium concentration pools, etc. It is used as the basis for defining the Voltage-GatedChannel, Ligand-Activated Channel, and IonicPump templates. At this time, we have only defined Voltage-GatedChannel; it is discussed in the next section.
![\includegraphics[scale = 0.65]{transmembrane.eps}](img22.gif)
The TransmembraneMechanism template inherits from Model the attributes name, description, notes, author, and reference. It adds three new attributes of its own: cellType, experimentalData, and pharmacodynamics. The cellType attribute is a string that allows users to describe the type of neural cell involved. It has a semi-controlled vocabulary associated with it listed in Figure 4. The user may select from one of the values or supply a different value of their choice. The attribute experimentalData is a list of pointers to Data objects, for referring to experimental data relevant to a particular model of a transmembrane mechanism. The attribute pharmacodynamics is a list of Pharmacodynamics objects used to record information about pharmacodynamics experiments involving the transmembrane mechanism.
|
cellType
|
One assumption needs to be stated at the outset. It is not possible to take a generalized channel representation such as what is presented here and translate it directly into something appropriate for a simulator such as GENESIS or NEURON without additional assumptions. We assume that the programs which translate channel representations into simulator code will have specific expectations about the form of the representation. We expect also that the translator program will augment the representations with additional elements that are not explicitly state. For example, a working simulation involving channels needs to have variables for quantities such as the channel current. Rather than try to build into the representation here something that lists which variables should be added to a simulation script, we assume that the translation program will construct simulator-specific code using both the channel model representation and additional code that is common to all channel models.
As its name implies, the Voltage-GatedChannel template can be used to represent voltage-gated ion channels. The representation is based in large part on that used by GENESIS, and can handle channels not only of the common Hodgkin-Huxley variety, but also a number of variants. The representation is fairly complex, so the following discussion is divided into parts.
![\includegraphics[scale = 0.65]{voltagegated.eps}](img23.gif)
The diagram for Voltage-GatedChannel shows that the top-level structure consists of a number of attributes and a list of ions. Attributes channelType and currentType are strings used to describe the type of channel and the type of current it passes. The user is free to choose any values for these attributes. An example value for the channelType attribute may be ``K'' (for potassium), whereas for currentType, it might be ``K(A)'', signifying an A-type transient K current. The attributes cellRestingMembranePotential and cellRestingMembranePotential_units record information about the resting membrane potential of the cell in which the channel is found. The attributes temperature, Q10Factor and scalingFactor specify the temperature assumed for the parameter values in the model and scaling factors to be used to adjust parameters for other temperatures if needed.
A given channel may pass more than one kind of ion, although current channel models most often involve only one ion. Nevertheless, to handle the case of multiple ions, the ion attribute in Voltage-GatedChannel is a list of Ion structures. Each Ion structure contains the following attributes: name, a string that records the user-chosen name of the ion (e.g., ``potassium''); isMainIon, a boolean flag that records whether the ion is the main one passed by a channel or whether it is a secondary ion (for those channels that involve multiple ions); equilibriumPotential, a floating point value representing the membrane potential at which there is no net flux of the ion across the membrane; and gatingVariable, a list of one to three GatingVariable structures.
In neuronal modeling, individual ion channels are often though of as containing a small number of gates that control the flow of ions through the channel. The GatingVariable structure contains attributes related to the properties of a gate. The collection of gating variables in a particular channel model captures the bulk of the information necessary to describe the behavior of the channel. Each GatingVariable structure contains the following attributes:
| (1) |
We have eschewed attempting to provide separate structures for every different variation on voltage-gated channels. Instead, the approach here uses a common model organization that maps all the variations into one of three types of representations: a parametrized form, a tabulated form, and a catch-all equation-form. Within the parametrized form there are several predefined common types. The three forms are subclasses of GatingVariableRepresentation, and are described below.
Each gating variable can be independently described using one of the three main forms. This gives the user freedom to model categories of channels that use combinations of, say, a gating variable described using a parametrized form and another gating variable described using a tabulated form. Regardless of the particular form, every gating variable has two rate functions, one for forward and one for backward. Each of the three subclasses of GatingVariableRepresentation therefore have two attributes, forward and backward.
The version of the Hodgkin-Huxley model of voltage-gated channels used here is based on a general format used in GENESIS. It is characterized by three main assumptions about the equations for channel conductance and the forward and backward rate functions. First, the model assumes an Ohm's law relationship between current and conductance, with
| (2) | |
| (3) |
| (4) | ||
| (5) | ||
| (6) |
The attribute category in the ParametrizedForm structure
determines which of several variations of this equation should be presented
to the user. We have so far determined the following categories: ``General
Parametrized Form'', ``Hodgkin-Huxley Exponential'', ``Hodgkin-Huxley
Sigmoid'', ``Hodgkin-Huxley Linoid'', ``Sigmoidal ![]() and constant
and constant
![]() '', and ``Borg-Graham''.
'', and ``Borg-Graham''.
Formula 7 given above is the most general; it is called the
``General Parametrized Form''. This same formula for ![]() and
and ![]() can represent the three common forms of Hodgkin-Huxley equations:
can represent the three common forms of Hodgkin-Huxley equations:
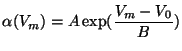 |
(8) |
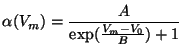 |
(9) |
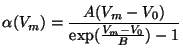 |
(10) |
We assume that the user interface for a tool such as Modeler's Workspace will provide a fill-in-the-blanks facility that triggers on the user's choice from a pull-down list for the category attribute, and displays one of the specialized equation forms for user input. This will help present the user with a slightly more familiar format, for those cases when they are interested in using one of the common types of equations.
ParametrizedForm does not have attributes related to tabular fill or interpolation. The reason is that ParametrizedForm is intended to describe gate variables using a particular, general equation. The issue of how the generalized equation is implemented in a simulator (for example, whether it is internally turned into a table of numbers, as in GENESIS' tabchannel) is a simulator-specific issue. Some simulators may not turn the generalized equation into a table at all. Therefore, it does not seem suitable to provide attributes related to table translation in the ParametrizedForm data structure. However, it is appropriate for the TabulatedForm, and therefore, TabulatedForm does have attributes that let a user specify how a table should be expanded by interpolation; see the next subsection.
The TabulatedForm structure is used to represent gate variables with tabulated data. This can be useful in cases where a modeler has experimental data characterizing the behavior of a channel gate. The representation here provides for separate tables for the forward and backward rate functions. Each can be a one- or two-dimensional table of floating-point quantities stored in elem, along with variables startingValue and endingValue that express the range of the independent quantity in the table. Both startingValue and endingValue are lists of one-to-two floating-point numbers, to handle the case of 1-D and 2-D tables.
The interpolation attribute, using the InterpolationParameters structure, allows a user to express expectations about how a given table should be expanded by interpolation. This is useful if the user has a small number of data points, but expects those data points to be used to fill a larger table using a particular interpolation method. The possible values of method are restricted to a small number of predefined methods, to help ensure users see the same results in different simulation programs. The attribute interpolatedSize
The EquationForm structure stores rate functions expressed as formulas. The Equation structure has the following parameters for this purpose: equation, a text string representing a formula; startingVoltage and endingVoltage, floating-point values representing the range of voltage values for which the formula is valid; assign, a list of ParameterAssignment structures for assigning values to symbolic parameters used in the formula; and an optional divisions attribute that can be used to specify the number of divisions into which the equation should be discretized over the range startingVoltage to endingVoltage.
The syntax of the expression language permitted in the equation
attribute is taken from the C and Java languages. The differences are that
we allow the caret symbol (^) as a short-form replacement for the
pow function for exponentiation, and add the function
nint, which is available in some UNIX C libraries but is not an
ISO C standard function. The functions permitted in the text string
representation of the formula are: cos, sin,
tan, sqr, sqrt, pow, log,
log10, exp, asin, acos, atan,
abs, nint, ceil and floor.
(`log' here is the natural logarithm.) The operators permitted in
the text string representation of the formula are summarized in
Table 1. All operators in formulae return floating-point
values. For boolean operators, 0 and 0.0 are interpreted as ``false'' and
all other values are interpreted as ``true''.
The conditional operator, ``? :'', warrants some elaboration. It
is sometimes useful to express a discontinuous rate function. For example,
Traub et al. (1991) use the following formula for ![]() , a forward
inactivation rate function used in one of their channel models:
, a forward
inactivation rate function used in one of their channel models:
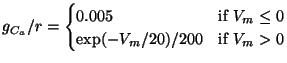 |
(11) |
Finally, another point is worth clarifying in this context. EquationForm is not intended to express an equation that is used to fill a TabulatedForm table. The three representational forms (ParametrizedForm, TabulatedForm, EquationForm) are independent methods for describing channel behavior. A GENESIS or NEURON simulator plugin translating an EquationForm representation may generate a script-language function that embodies the EquationForm equation, or it may create a discretized, tabulated representation of the funtion. The latter case would be likely for GENESIS. However, if the translation program does produce a tabular representation of the equation, it should not then store the results in a TabulatedForm representation in the model definition. The different forms store the user's input; they should not be modified by a simulator plugin translation program.
To implement certain kinds of channels in a simulation, certain quantities must be exchanged between the channel gating variables and other elements in the cell model within which the channel is situated. This kind of value-passing or message-passing is specified using the ModelMessage structure list attached to the message attribute in GatingVariable.
A given Voltage-GatedChannel object may have zero or more messages. Each ModelMessage structure specifies a message name (e.g., ``voltage''), a source or destination (which could be, e.g., a calcium concentration object created from a ConcentrationPool template), and a direction (i.e., whether the message is coming from a source or going to a destination). References to models are implemented using XML XLink, with two attributes source and destination, both pointing to Model objects. This allows linking to any database object that is derived from the Model template, including all Neuron and channel models.
The IntracellularMechanism template is intended to serve as a common starting point for defining items such as calcium concentration pools.
This section is currently unfinished.
In this section, we present an example of translating a model into the Modeler's Workspace representation defined in this document. The approach to translating into XML Schemas the object class definitions presented in the sections above is described in the companion document The Modeler's Workspace Notation for Describing Representations Intended for XML Encoding (Hucka, 2000). Appendix B gives the full listing of a preliminary version of an XML Schema corresponding to the Modeler's Workspace representation.
The model of a hippocampal pyramidal cell by Traub et al. (1991) requires defining several channel objects and an overall cell object. The channel models are listed below; the cell model is given in Section 6.2.

The object defined in this section is a rendition of the CA3 pyramidal neuron modeled by Traub et al. (1991). It refers to the channels defined in the previous section.
This section is currently unfinished.
The model described in this section refers to a number of external objects, such as Author and Reference. Example definitions are shown below.





The Modeler's Workspace Database uses a unique identifier scheme patterned after message-id's used in Internet electronic mail (Crocker, 1982; Färber, 1998; Levinson, 1997). The MWSUID data type in Modeler's Workspace representations stores such an identifier. The technique for generating a MWSUID value is fairly simple, and is based on concatenating the following components, in order, into a text string:
The value of the ``current time'' used above is taken as the first time the object is saved by the user to the Workspace Database. Thereafter, the time component of the identifier is never changed--the object identifier is left frozen for that database object. If the object is edited, it retains its unique identifer; however, if the object is copied with the same database, or deleted, the unique identifier is lost and not reused. New objects or object copies always receive a new identifier. An example of a unique identifier generated using this scheme is: ``1484450292207948AF2C@obi-wan.bbb.caltech.edu''.
This section is currently unfinished.
This document was generated using the LaTeX2HTML translator Version 99.2beta8 (1.46)
Copyright © 1993, 1994, 1995, 1996,
Nikos Drakos,
Computer Based Learning Unit, University of Leeds.
Copyright © 1997, 1998, 1999,
Ross Moore,
Mathematics Department, Macquarie University, Sydney.
The command line arguments were:
latex2html -white -split +0 -show_section_numbers -image_type gif -no_navigation -local_icons -discard -mkdir -dir html mws-rep
The translation was initiated by on 2001-06-21