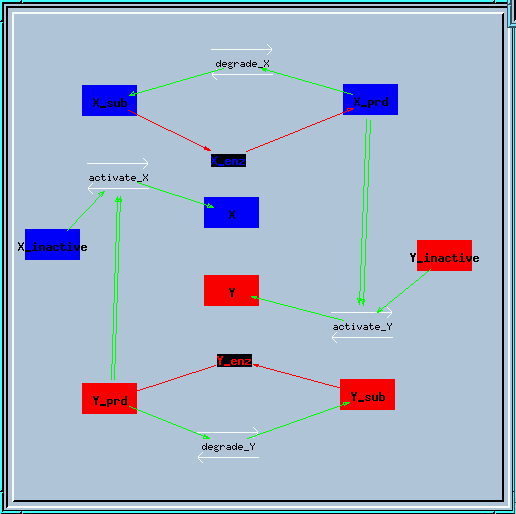
Bistable feedback loop reaction diagram
Kinetikit, developed by Upinder S. Bhalla (National Centre for Biological Sciences, Bangalore, India), is a utility and graphical interface for modeling large numbers of chemical reactions such as occur in biochemical signaling pathways. It is designed to be 'click and drag' and provides a user-friendly interface to the GENESIS kinetics modeling library functions for both research and educational uses. Its use is described in the online help for Kinetikit, and in detail in Chapter 10 of the Book of GENESIS. Further research applications are described in Bhalla and Iyengar (1999).
The example below was taken from the Kinetkit page at http://www.ncbs.res.in It describes the example Kinetikit simulaton given in the GENESIS directory Scripts/kinetikit/examples/feedback.g.

In this feedback loop, the enzyme X creates a product X_prd, which activates enzyme Y in a second-order manner. Similarly, enzyme Y produces Y_prd to activate X.
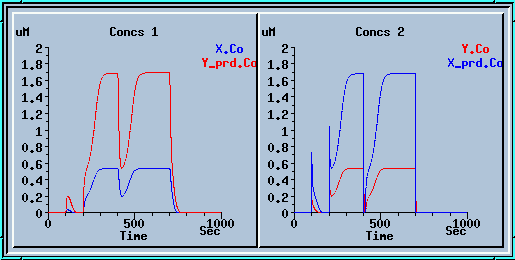
This is a plot of the output of the feedback loop. An explanation of the output and further examples of Kinetikit simulations can be found at the Kinetkit home page.
 Return to the summary of tutorials
Return to the summary of tutorials