


Next: IMPLEMENTATION OF THE
Up: The GENESIS Simulator-based Neuronal
Previous: GENESIS AS A
We have decided, based on the current design of the database system, to
focus the development of the database prototype specifically on the models we
have constructed related to the mammalian cerebellum. We have determined
that this specific focus will allow us to fully develop a working prototype
of the database system. At present, several models exist at the single cell
level, which will form the basis for the database. Over the last year, we
have completed construction of a systems level-network model within GENESIS
which includes major projections to cerebellar folium, crus IIa. The model
incorporates physiological data on the structure of cerebellar tactile maps
as well as maps for the trigeminal nuclei (Principalis and Interpolaris),
thalamic nuclei (VPM and POM) and somatosensory cortex. This later data was
obtained from the literature. Over the last year, we have also begun to
develop a network model of cerebellar cortex. This large project will take
several years to complete, but will, when finished allow us to link the
systems level model to the single cell models currently complete or being
constructed. From the point of view of the database, linking these models
together will provide a prototype for a multi-level browsing interface of
neurobiological data relevant to the functional organization of the
cerebellar cortex.
The following example illustrates the way in which the database would
be used. In this example, we assume that the interface to the multibase
system has been implemented on a high-end graphics workstation where a
windowing environment and mouse is a standard means for interaction. Let us
assume that a neuroscientist is interested in exploring ways in which
calcium channels affect the firing of Purkinje cells in the cerebellum.
Figure 1 shows a prototype for the top level interface to the database.
After entering the keyword ``purkinje'', the user is presented with a number
of contexts in which Purkinje cells appear in the database, grouped into
several categories. From left to right, the categories are (a) behavioral
studies (of tactile exploration and cerebellar lesions), (b) the systems
level (in this case, what is known about the connections between regions of
the cerebellar system), (c) the network level (a model of the cerebellar
cortex), (d) the cellular level (Purkinje cell model), (e) the subcellular
level (ion channels, etc.), and (f) pertinent references to the literature.
These various categories may be accessed both from the top level interface
(the Multi-Browser), and from within other categories.
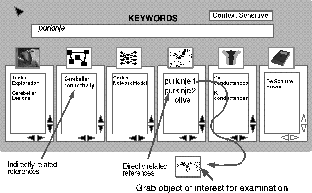
Figure 1: The prototype for the top level interface to the database.
Several levels of contexts for the keyword "purkinje" are displayed,
ranging from behavioral studies of the animal to subcellular properties
of the Purkinje cell, plus pertinent references to the literature.
The selection of the De Schutter and Bower (1994a,b) Purkinje cell model
leads to a tutorial on the model, with the opportunity to view a description
of the model and simulation results of voltage clamp, current injection,
synaptic input, and simulated in vivo responses. (See
COLOR PLATE.)
Click HERE to see the large color figure.
COLOR PLATE:
A tutorial on the cerebellar Purkinje cell is used as an entry point to
queries on Purkinje cells, when using the GENESIS Simulator-based Neuronal
Database. Here, the user has selected the Voltage Clamp tutorial. In
addition to showing both simulated and experimental results for voltage
clamp results, this tutorial provides a list of suitable keywords for
additional searches. For example, "channels" will allow the the user to
extract information regarding the voltage dependent channels which are found
in Purkinje cells. The detailed cell model (De Schutter & Bower 1994a,b)
is showing a false color representation of the membrane potential throughout
the cell, as a result of a previously performed current clamp simulation.
Each of these categories allow an examination of the experimental data for
comparison. From within the tutorial, as well as from the subcellular
category of the top level interface, the object shown in Figure 2 may be
accessed. Here, it is possible to query the model for the parameters used
to characterize each of the ten types of channels used in the model (lower
left), the distribution of these conductances within the cell model, and
references to the papers which provided the descriptions of the channels
which were implemented in the model.
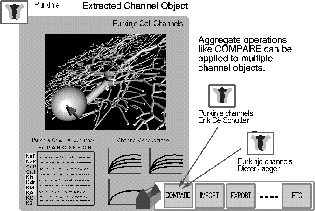
Figure 2: The tutorial on Purkinje cell channels, accessible from either the
top level
interface or the Purkinje cell tutorial, provides a basis for queries
regarding channel properties. The detailed cell model shown (De Schutter &
Bower 1994a,b) uses 4550 compartments to model the dendrites and dendritic
spines, and contains 8021 ion channels which fall into 10 categories. The
tool box at the lower left provides a way to import and export other data,
and to make comparisions with other objects in the database.
The tool box at the lower left provides a way to import and export other
data. As it is important to understand the basis of assumptions which were
made in the model, there is access to the experimental current injection
results which were used to ``tune'' the model parameters. Links to remote
sites are provided so that it is possible to access the full data set, as
well as the data used for tuning. Objects from the database may be exported
to GENESIS simulations, data analysis tools, and visualization tools. The
COMPARE button provides tools for aggregate operations such as making
automated comparisions with other objects in the database. For example,
comparisons may be made with channels found in another model. Within the
tutorial it will be possible to perform searches to answer questions like
``what are all the single-cell models which use this type of Ca channel with
parameters in this range; what are all the bursting models that were based
on spiking behaviour with these statistics?" The researcher can also move
from a Ca channel in a Purkinje cell to a listing of all the places Ca
channels are currently used in models in GENESIS.
At this point, the user elects to view predictions of in vivo physiological
responses to real stimuli, bringing up the display shown in Figure 3.
The link at the lower left allows the viewing of experimental extracellular
recordings and peri-stimulus time (PST) histograms for comparison with the
simulated results. By clicking on a set of results, it is possible to
find a reference for the source of these results (De Schutter & Bower,
1994b).
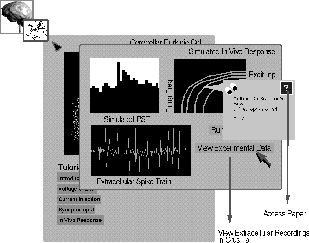
Figure 3: Predictions of in vivo physiological responses to appied stimuli are
accessed from the Purkinje cell tutorial. Both simulated and experimentally
obtained measurements of PST histograms, spike trains, and other data may
be viewed.
Having explored the single cell level, the user may switch to the cerebellar
network level and explore the response of the model of the cerebellar cortex
(crus IIa) of the rat to selected input patterns of stimulus. Figure 4
shows the result of a query for the corresponding experimental results.
Here, the cursor is used to select tactile stimulation applied to the
ipsilateral upper lip area (IUL) in the diagram at the upper right. This
results in a display of the PST histogram obtained from recordings from the
point marked ``X'' in crus IIa, which is within the patch that is
somatopically mapped to the IUL. The peaks in the PST histogram correspond
to information coming from two different pathways - the direct pathway
through the trigeminal nucleus, and the indirect pathway from the
somatosensory cortex. A query for the source of the experimental data leads
to a reference to the work of Thompson and Bower (1993).
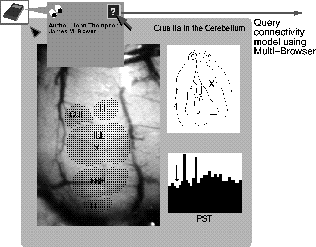
Figure 4: The result of a query for experimental results showing the response
of the
cerebellar cortex of a rat to facial stimulation. The ipsilateral upper lip
area (IUL) was selected, and the PST histogram displayed is for recordings
at the corresponding somatopically mapped area of crus IIa.
Moving up another level, the user then shifts to the cerebellar system model
in order to learn more about the pathways which are involved. Figure 5
shows a simplified representation of the major sensory pathways to the
cerebellum. This is an interface to the high level GENESIS systems level
model of the cerebellum and the regions projecting to it. This model lets
one click on particular regions of either somatosensory cortex, the
thalamus, the trigeminal nucleus, the cerebellum, the pons, or the animal's
face, and see all the related regions in each structure. It is possible to
see where they connect as well as what their receptive fields are. One can
also click on different buttons and see the secondary receptive fields.
This can be done in the normal animal simulation as well as in the
simulation of an animal with lesioned trigeminal nerve. When the trigeminal
nerve is cut, these original projections are kept constant and the model may
be used to test the hypothesis that the reorganization that is observed
experimentally in the cerebellum could happen without any new projections.
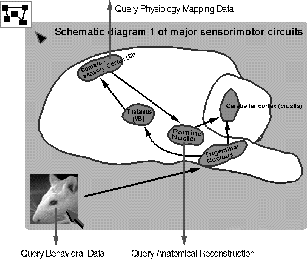
Figure 5: The graphical interface for queries regarding Purkinje cells at the
systems
level. Selecting particular regions in the diagram of the major sensory
pathways to the cerebellum allows one to see both simulated and experimental
data for primary and secondary receptive fields under various conditions.
The projections are all established based on the reports in the literature
of the receptive field size of each different brain area. It is possible to
query this data, as well as descriptions of the experimental procedures. By
clicking at the lower left of Figure 5, it is possible to query the highest
level of the database, and examine data for the behavior of the rat before
and after severing the trigeminal nerve.
The user can repeat all of the above for the other models found in the
original query in order to make comparisons between them. With all this
information available, the researcher can develop a better understanding of
the difference between the models as well as their similarities. If the
neuroscientist is interested in developing a new model or exploring
different mechanisms, then the database will provide information on which
sets of parameters are the most appropriate to use in the simulations that
will be performed.



Next: IMPLEMENTATION OF THE
Up: The GENESIS Simulator-based Neuronal
Previous: GENESIS AS A
Dave Beeman
Wed Oct 11 15:28:38 PDT 1995




